NYU iGEM: Biosensors for Water Contamination, an NYU startup led by Ali Hasan (Tandon BS ‘20) and David Sung (Tandon BS ‘21), share lessons learned through their participation in the Prototyping Fund.
We are Ali Hasan (Tandon ‘20) and David Sung (Tandon ‘21), members of the NYU iGEM team. iGEM (International Genetic Engineering Machine) is an organization dedicated to solving global issues in various fields- including agricultural, environmental, and pharmaceutical- using synthetic biology. Every year, iGEM teams from different universities around the world present their work at the iGEM Synthetic Biology conference hosted in Boston, Massachusetts.
Phase 1- Last year, our project focused on the sustainable production of flavonoids, a family of compounds commonly found in plants and known to have pharmaceutical properties. Unlike conventional chemical synthesis or extraction from plants, which methods are very expensive and environmentally harming, synthetic biology offers a more viable and sustainable approach to pharmaceutical production.
Through genetic engineering, we were able to program bacteria to produce various flavonoid compounds. In addition, we implemented a novel technique called optogenetics. In the presence of different colored lights, the bacteria produces a different flavonoid compound. With this approach, we are able to manufacture a specific drug compound depending on our needs.
In order to mass-produce the drugs, we had to design a lab-scale bioreactor that would provide an optical environment for the bacteria to grow. Thus, our design included equipping our bioreactor with temperature control, oxygen feed, O2 sensor, pH sensor, and most importantly the LED light system.
From the Phase 1 funding we received from the Prototyping Fund, we were able to make our vision into reality and assemble a fully-functioning bioreactor. At the 2019 iGEM conference, we showcased our optogenetic project and the final bioreactor design and won a Silver Medal at the conference!
Phase 2-
After presenting at the iGEM conference, we began to brainstorm ideas for a new project. We looked at real-world problems that we may be able to solve or deal with using synthetic biology. We found that water contamination was a big problem here in the United States as well as in developing nations. It’s estimated that a quarter of Americans drink water from systems that don’t meet safety standards. This makes sense given that the American Society of Civil Engineers gives US infrastructure a D+ rating. One of the most notable examples of water contamination in the US is the crisis in Flint, Michigan. Contaminants can include lead, arsenic, rust, and even chloroform. Home testing devices are often inaccurate and usually only test for a single contaminant. Water samples need to be sent to special water labs to accurately test for contamination. For this year’s project, we decided on genetically engineering bacteria to act as biosensors for contaminants in water.
Using our previous project as our inspiration, we began designing our water contaminant sensor. However, unlike before when we were focusing on large-scale production of pharmaceutical compounds, our goal now was to detect contaminants in the scale of parts per million. What we needed was not a large bioreactor, but a scaled-down biosensor that would fit on the palms of our hand. What we needed was a microfluidic “lab-on-a chip” device. A microfluidic platform would provide us with a cheap and consistent way to test water.
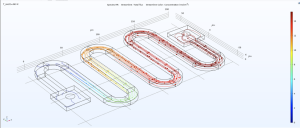
Modeling the Microfluidic Chip
Using COMSOL, a powerful simulation software, we have been able to model our microfluidic chips.
Modelling our device through COMSOL will show us how the device will function and help us optimize the design of the chip. We will continue increasing the complexity of the model over the summer so we can gather more information and are ready to build our device once the school opens up again.